MAG Generation
MAG generation
- Generation of metagenome assembled genomes (MAGs) from assemblies
- Assessment of quality
- Taxonomic assignment
Prerequisites
For this tutorial, you will need to first start the docker container by running:
sudo docker run --rm -it -v /home/training/Binning:/opt/data microbiomeinformatics/mgnify-ebi-2020-binningpassword: training
Generating metagenome assembled genomes
Learning Objectives - in the following exercises, you will learn how to bin an assembly, assess the quality of this assembly with checkM, and then visualize a placement of these genomes within a reference tree.
As with the assembly process, there are many software tools available for binning metagenome assemblies. Examples include, but are not limited to:
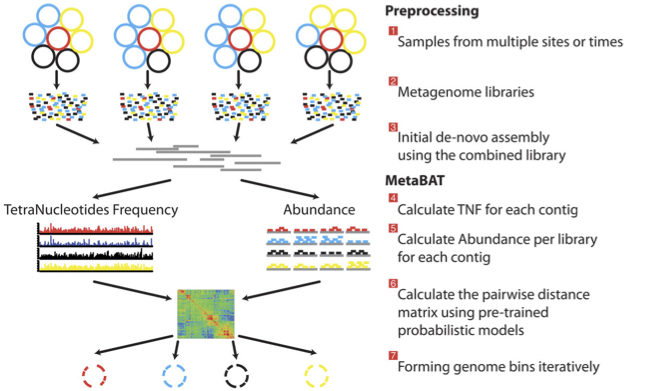
There is no clear winner between these tools, so it is best to experiment and compare a few different ones to determine which works best for your dataset. For this exercise, we will be using MetaBAT (specifically, MetaBAT2). The way in which MetaBAT bins contigs together is summarized in Figure 1.
Preparing to run MetaBAT
If you wanted to generate the input.fastq.sam.bam file yourself, you would run the following commands:
# NOTE: you will not be able to run subsequent steps until this workflow is completed because you need
# the input.fastq.sam.bam file to calculate contig depth in the next step.
# If you would like to practice this now, back up the input.fastq.sam.bam file that we provided first,
# as these steps will take a while:
cd /opt/data/assemblies/
mv input.fastq.sam.bam input.fastq.sam.bam.bak
# index the contigs file that was produced by metaSPAdes:
bwa index contigs.fasta
# fetch the reads from ENA
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/ERR011/ERR011322/ERR011322_1.fastq.gz
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/ERR011/ERR011322/ERR011322_2.fastq.gz
# map the original reads to the contigs:
bwa mem contigs.fasta ERR011322_1.fastq ERR011322_2.fastq > input.fastq.sam
# reformat the file with samtools:
samtools view -Sbu input.fastq.sam > junk
samtools sort junk -o input.fastq.sam.bamRunning MetaBAT
In this case, the directory might already be part of your VM, so do not worry if you get an error saying the directory already exists. You can move on to the next step.
Once the binning process is complete, each bin will be grouped into a multi-fasta file with a name structure of bin.[0-9].fa.
Obviously, not all bins will have the same level of accuracy since some might represent a very small fraction of a potential species present in your dataset. To further assess the quality of the bins, we will use CheckM.
Running CheckM
CheckM has its own reference database of single-copy marker genes. Essentially, based on the proportion of these markers detected in the bin, the number of copies of each, and how different they are, it will determine the level of completeness, contamination, and strain heterogeneity of the predicted genome.
This program has some handy tools not only for quality control but also for taxonomic classification, assessing coverage, building a phylogenetic tree, etc. The most relevant ones for this exercise are wrapped into the lineage_wf workflow.
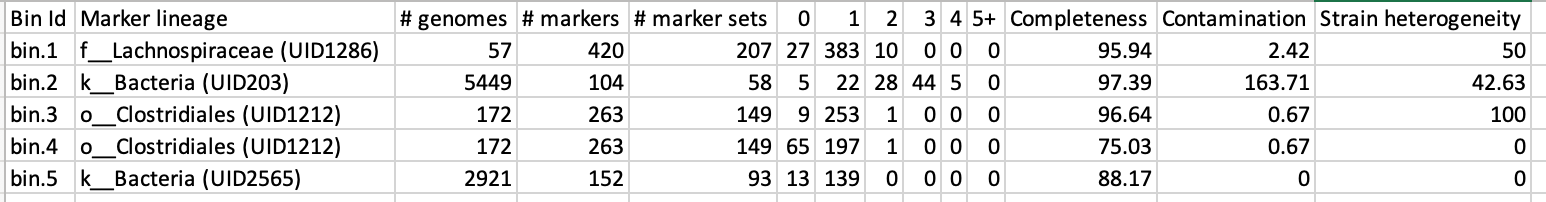
Before we can visualize and plot the tree, we will need to convert the reference ID names used by CheckM to taxon names. We have already prepared a mapping file for renaming the tree (rename_list.tab). We can then do this easily with the newick utilities.
Visualizing the phylogenetic tree
We will now plot and visualize the tree we have produced. A quick and user-friendly way to do this is to use the web-based interactive Tree of Life iTOL
iTOL only takes in newick formatted trees, so we need to quickly reformat the tree with FigTree.
Go to the iTOL website (open the link in a new window).
The login is as follows:
User: EBI_training
Password: EBI_training
After you login, just click on My Trees in the toolbar at the top and select
IBD_checkm.nwk from the Imported trees workspace.
Alternatively, if you want to create your own account and plot the tree yourself follow these steps:
- After you have created and logged in to your account go to My Trees
- From there select Upload tree files and upload the tree you exported from FigTree
- Once uploaded, click the tree name to visualize the plot
- To colour the clades and the outside circle according to the phylum of each strain, drag and drop the files iTOL_clades.txt and iTOL_ocircles.txt present in /home/training/Data/Binning/iTOL_Files/ into the browser window
Once that is done, all the reference genomes used by CheckM will be coloured according to their phylum name, while all the other ones left blank correspond to the target genomes we placed in the tree. Highlighting each tip of the phylogeny will let you see the whole taxon/sample name. Feel free to play around with the plot.
To find the bins you generated, click on the search icon in the left-hand side menu (magnifying glass with the letters “Aa” in it). In the search field type bin.. Click on each bin name to see it in the tree.